Hands-On Exercise 8: browse published results¶
In this exercise, we will run again the workflow of the exercise a multi-node workflow, then we will browse its results through the Oozie Web Console that was previously introduced.
Prerequisites¶
- You have cloned the Hands-On git repository (see Clone the Hands-On repository),
- (Only for python) You have installed the required software (see Prerequisites when using python).
Install the Hands-On¶
- Install the Hands-On Exercise 8, just type:
cd
cd dcs-hands-on
mvn clean install -D hands.on=8 -P bash
Run the workflow¶
- Type the following command:
ciop-run
- Wait until the workflow is completed (it will take approximately five minutes).
Browse the results¶
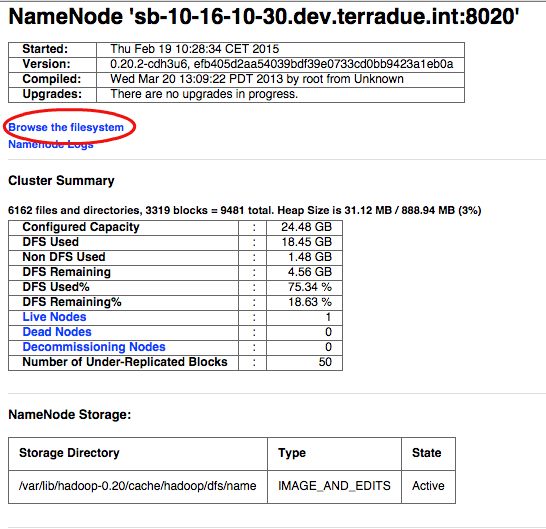
- Open a browser and type http://$HOSTNAME:50070 ,
Tip
Read how to obtain the value of your Sandbox $HOSTNAME in the General Notes section
- Click on the link Browse the filesystem,
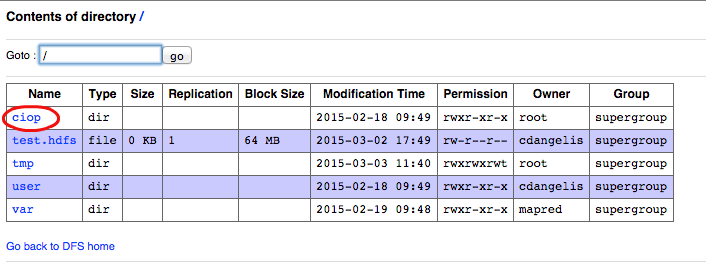
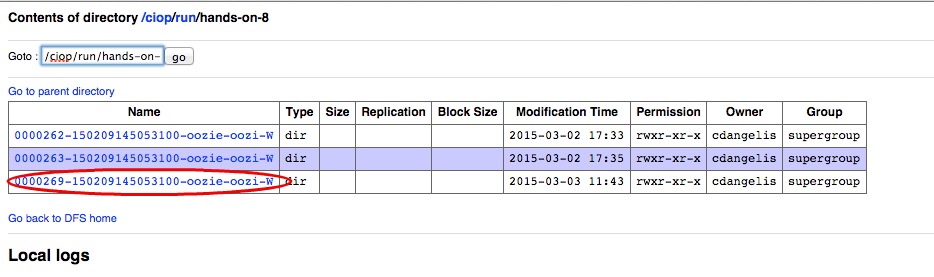
- Click on the link ciop,
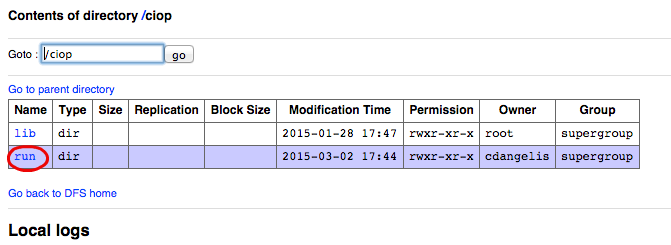
- Click on the link run,
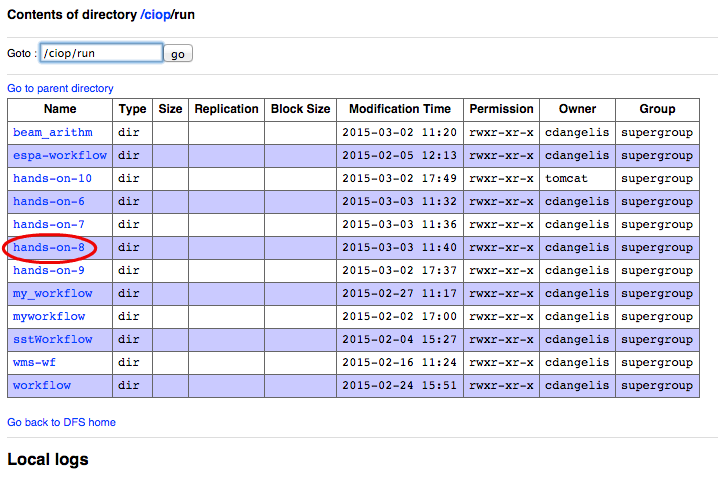
- Click on the link hands-on-8,
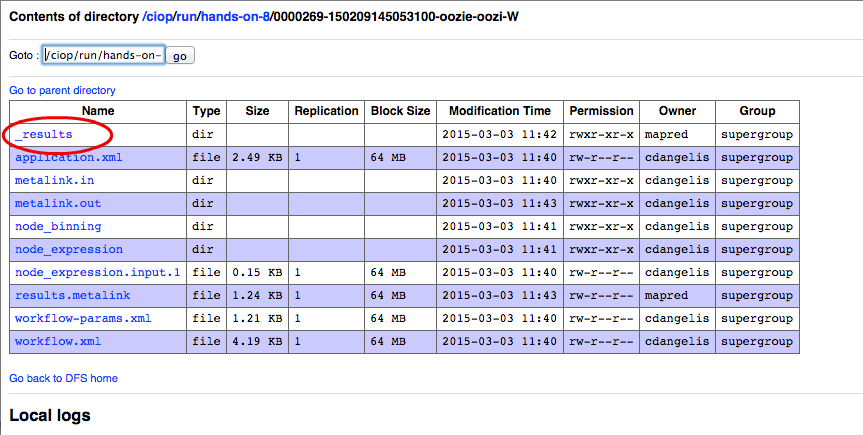
- Click on the link representing the workflow id (e.g., 0000269-150209145053100-oozie-oozi-W),
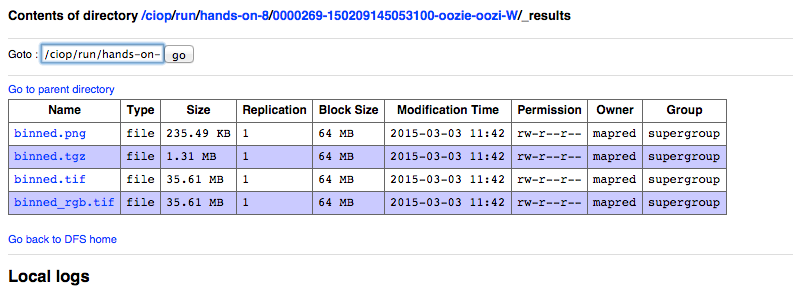
- Click on the link _result,
- To see intermediate results, click on node_expression and then click on data.
The following images show the steps that we have just performed:
Access the results from a shell¶
You can also access the results from a shell or from scripts. Since the results are on the HDFS file system, you have to use the hadoop command with its subcommand fs.
For example, to see the results from the example above, you would have to use the following command:
hadoop fs -ls /ciop/run/hands-on-8/0000269-150209145053100/_results
You can use other shell-like subcommands to access, manipulate and transfer files. For a full guide of HDFS commands, see the official documentation on the Apache web site.
Recap¶
- We executed a multinode workflow;
- We browsed either final or intermediate results through the Oozie Web Console.