better-satcen-00001 data pipeline results (Sentinel-2 Vegetation and Water themtic index) : loading from pickle file and exploiting¶
This Notebook shows how to: * reload better-satcen-00001 data pipeline results previously stored in a pickle file * Create a new dataframe with data values cropped over the area of interest * Plot an RGB * Create a geotif
Import needed modules¶
In [1]:
import pandas as pd
from geopandas import GeoDataFrame
import gdal
import numpy as np
from shapely.geometry import Point
from shapely.geometry import Polygon
import matplotlib
import matplotlib.pyplot as plt
from PIL import Image
%matplotlib inline
from shapely.wkt import loads
from shapely.geometry import box
from urlparse import urlparse
import requests
Read picke data file¶
In [2]:
pickle_filename = 'better-satcen-00001_May_2018_Albania-GreeceBorder.pkl'
In [3]:
results = GeoDataFrame(pd.read_pickle(pickle_filename))
In [4]:
results.head()
Out[4]:
| enclosure | enddate | identifier | self | startdate | title | wkt | |
|---|---|---|---|---|---|---|---|
| 0 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 0712a0fdc17bad31138dfef3f6d57a6cb66962ea | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TEK_2... | POLYGON((20.9997634361085 39.661229210097,21.2... |
| 1 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 07fb7271ad6b5c5d2b23ff305c617c0a532926bc | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TEK_2... | POLYGON((20.9997634361085 39.661229210097,21.2... |
| 2 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 15412a409b5f269b90a6c4edd71a70355830ed6f | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_2... | POLYGON((19.8899808364281 40.5626923867771,21.... |
| 3 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 238107f531ed150ebb3ee0763d3d690ec791b305 | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TEK_2... | POLYGON((20.9997634361085 39.661229210097,21.2... |
| 4 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 2eebf44a805a2de437e839a6245260e6c2449948 | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TEK_2... | POLYGON((20.9997634361085 39.661229210097,21.2... |
Credentials for ellip platform access¶
- Provide here your ellip username and api key for platform access
In [5]:
import getpass
user = getpass.getpass("Enter your username:")
In [6]:
api_key = getpass.getpass("Enter the API key:")
Define filter parameters¶
Time of interest¶
In [7]:
mydates = ['2018-05-22 09:30:29.027','2018-05-14 09:20:31.024000', '2018-05-12 09:30:29.027000000', '2018-05-09 09:20:29.027000', '2018-05-07 09:30:41.024000', '2018-05-02 09:30:39.027000']
Area of interest¶
- The user can choose to define an AOI selecting a Point and a buffer size used to build a squared polygon around that point
In [8]:
point_of_interest = Point(20.951, 40.768)
In [9]:
buffer_size = 0.07
In [10]:
aoi_wkt1 = box(*point_of_interest.buffer(buffer_size).bounds)
In [11]:
aoi_wkt1.wkt
Out[11]:
'POLYGON ((21.021 40.698, 21.021 40.838, 20.881 40.838, 20.881 40.698, 21.021 40.698))'
- Or creating a Polygon from a points list (in this case this is exactly the Albania/Greece border reference AOI)
In [12]:
aoi_wkt2 = Polygon([(21.2961, 39.58638), (21.2961, 41.032), (19.8978, 41.032), (19.8978, 39.58638), (21.2961, 39.58638)])
In [13]:
aoi_wkt2.wkt
Out[13]:
'POLYGON ((21.2961 39.58638, 21.2961 41.032, 19.8978 41.032, 19.8978 39.58638, 21.2961 39.58638))'
Create a new dataframe with data values¶
- Get the subframe data values related to selected tiles and bands, and create a new GeoDataFrame having the original metadata stack with the specific bands data extended info (data values, size, and geo projection and transformation).
Define auxiliary methods to create new dataframe¶
- The first method gets tile reference, bands numbers, cropping flag, AOI cropping area and returns the related n bands data array with related geospatial infos
- If crop = False the original bands extent is returned
In [14]:
def get_bands_as_array(url,crop,bbox):
#[-projwin ulx uly lrx lry]
ulx, uly, lrx, lry = bbox[0], bbox[3], bbox[2], bbox[1]
output = '/vsimem/clip.tif'
ds = gdal.Open('/vsicurl/%s' % url)
ds = gdal.Translate(output, ds, projWin = [ulx, uly, lrx, lry], projWinSRS = 'EPSG:4326')
ds = None
ds = gdal.Open('/vsimem/clip.tif')
band = ds.GetRasterBand(1)
w = band.XSize
h = band.YSize
geo_transform = ds.GetGeoTransform()
projection = ds.GetProjection()
data = band.ReadAsArray(0, 0, w, h).astype(np.float)
data[data==-9999] = np.nan
ds = None
band = None
return data,geo_transform,projection,w,h
- The second one selects the data to be managed (only the actual result related to the selected tile) and returns a new GeoDataFrame containing the requested bands extended info with the original metadata set
In [15]:
def get_info(row, user, api_key, crop=False, bbox=None):
print 'Getting band %s' %(row['title'])
band_name = row['title'].rsplit('CROP_', 1)[1].split('.')[0]
parsed_url = urlparse(row['enclosure'])
url = '%s://%s:%s@%s/api%s' % (list(parsed_url)[0], user, api_key, list(parsed_url)[1], list(parsed_url)[2])
bands,geo_transform,projection,w,h = get_bands_as_array(url,crop,bbox)
extended_info = dict()
extended_info['band'] = band_name
extended_info['data'] = bands
extended_info['geo_transform'] = geo_transform
extended_info['projection'] = projection
extended_info['xsize'] = w
extended_info['ysize'] = h
series = pd.Series(extended_info)
print 'Get data values for %s : DONE' %row['title']
return series
- Define the reference tiles
In [16]:
ref_tiles = ['T34TDK', 'T34TEK', 'T34TDL']
- Define the selected bands (among the available ones) to be exploited
In [17]:
available_bands = ['ndwi', 'ndvi', 'quality_cloud_confidence', 'quality_snow_confidence', 'quality_scene_classification', 'B2', 'B3', 'B4', 'B5', 'B6', 'B7', 'B8', 'B8A', 'B11', 'B12']
selected_bands = ['ndwi', 'ndvi','B2','B3','B4']
- Select from metadataframe a subframe related to
- a single datetime
- multiple georeferenced tiles of interest
- selected bands
In [18]:
selected_bands = ['ndwi', 'ndvi','B2','B3','B4']
mydate_results = results[(results['startdate'] == mydates[0]) &
(results.apply(lambda row: row['title'].rsplit('CROP_', 1)[1].split('.')[0] in selected_bands, axis=1)) &
(results.apply(lambda row: ref_tiles[2] in row['title'], axis=1))]
mydate_results = mydate_results.merge(mydate_results.apply(lambda row: get_info(row, user, api_key, True, list(aoi_wkt1.bounds)), axis=1), left_index=True, right_index=True)
Getting band S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_ndwi.tif
Get data values for S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_ndwi.tif : DONE
Getting band S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_ndvi.tif
Get data values for S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_ndvi.tif : DONE
Getting band S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_B4.tif
Get data values for S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_B4.tif : DONE
Getting band S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_B2.tif
Get data values for S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_B2.tif : DONE
Getting band S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_B3.tif
Get data values for S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_B3.tif : DONE
In [19]:
mydate_results.head()
Out[19]:
| enclosure | enddate | identifier | self | startdate | title | wkt | band | data | geo_transform | projection | xsize | ysize | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 568c349b9dc75551e4ce5bfcd78331bbe836b3e5 | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_2... | POLYGON((19.8899808364281 40.5626923867771,21.... | ndwi | [[10850.0, 10492.0, 10808.0, 10545.0, 11087.0,... | (489960.0, 10.0, 0.0, 4520790.0, 0.0, -10.0) | PROJCS["WGS 84 / UTM zone 34N",GEOGCS["WGS 84"... | 1181 | 1555 |
| 18 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | f98960e9502a48c0e828c09286607409e34b910b | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_2... | POLYGON((19.8899808364281 40.5626923867771,21.... | ndvi | [[11299.0, 11599.0, 12047.0, 12331.0, 12509.0,... | (489960.0, 10.0, 0.0, 4520790.0, 0.0, -10.0) | PROJCS["WGS 84 / UTM zone 34N",GEOGCS["WGS 84"... | 1181 | 1555 |
| 26 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 5a40bae355f9fea6c4122adf6e0e97a202235689 | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_2... | POLYGON((19.8899808364281 40.5626923867771,21.... | B4 | [[4188.0, 3665.0, 3079.0, 2751.0, 2661.0, 2773... | (489960.0, 10.0, 0.0, 4520790.0, 0.0, -10.0) | PROJCS["WGS 84 / UTM zone 34N",GEOGCS["WGS 84"... | 1181 | 1555 |
| 27 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 661b503be3240860f0a99975f61a70ad255597b9 | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_2... | POLYGON((19.8899808364281 40.5626923867771,21.... | B2 | [[4241.0, 3696.0, 3080.0, 2722.0, 2758.0, 2684... | (489960.0, 10.0, 0.0, 4520790.0, 0.0, -10.0) | PROJCS["WGS 84 / UTM zone 34N",GEOGCS["WGS 84"... | 1181 | 1555 |
| 31 | https://store.terradue.com/better-satcen-00001... | 2018-05-22 09:30:29.027 | 7f6a9e0fac87968d4d090d1279a7f008b1ee88a1 | https://catalog.terradue.com//better-satcen-00... | 2018-05-22 09:30:29.027 | S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_2... | POLYGON((19.8899808364281 40.5626923867771,21.... | B3 | [[4328.0, 3711.0, 3236.0, 2829.0, 2795.0, 2848... | (489960.0, 10.0, 0.0, 4520790.0, 0.0, -10.0) | PROJCS["WGS 84 / UTM zone 34N",GEOGCS["WGS 84"... | 1181 | 1555 |
*Or*
- Select from metadataframe a subframe related ndvi and ndwi of different tiles in the same date:
- Show data values of NDVI bands
In [20]:
mydate_results[mydate_results['band'] == 'ndvi']['data'].values
Out[20]:
array([ array([[ 11299., 11599., 12047., ..., 9400., 9345., 9732.],
[ 11615., 11920., 12372., ..., 9732., 9411., 9727.],
[ 11706., 11853., 12393., ..., 9686., 9816., 9953.],
...,
[ 15029., 14047., 15687., ..., 16792., 17198., 17284.],
[ 15227., 13962., 16377., ..., 16928., 17042., 17412.],
[ 15799., 14860., 16004., ..., 17046., 16956., 17106.]])], dtype=object)
Plotting data and exporting geotif¶
Plot data¶
Plot all the selected bands
In [21]:
bands = None
bands = mydate_results['data'].values
titles = mydate_results['title'].values
numbands = len(bands)
print numbands
for i in range(numbands):
bmin = bands[i].min()
bmax = bands[i].max()
if bmin != bmax:
bands[i] = (bands[i] - bmin)/(bmax - bmin) * 255
else:
print 'WARNING bmin %s = bmax %s (%i)!' %(bmin, bmax,i)
5
- Plot the selected bands separately
In [22]:
fig = plt.figure(figsize=(20,20))
fig.suptitle('Results @ %s' % (mydates[0]), fontsize=16)
for i in range(numbands):
a = fig.add_subplot(3, 2, i+1)
#
imgplot = plt.imshow(bands[i].astype(np.uint8),
cmap='gray')
a.set_title(titles[i])
plt.tight_layout()
fig = plt.gcf()
plt.show()
fig.clf()
plt.close()
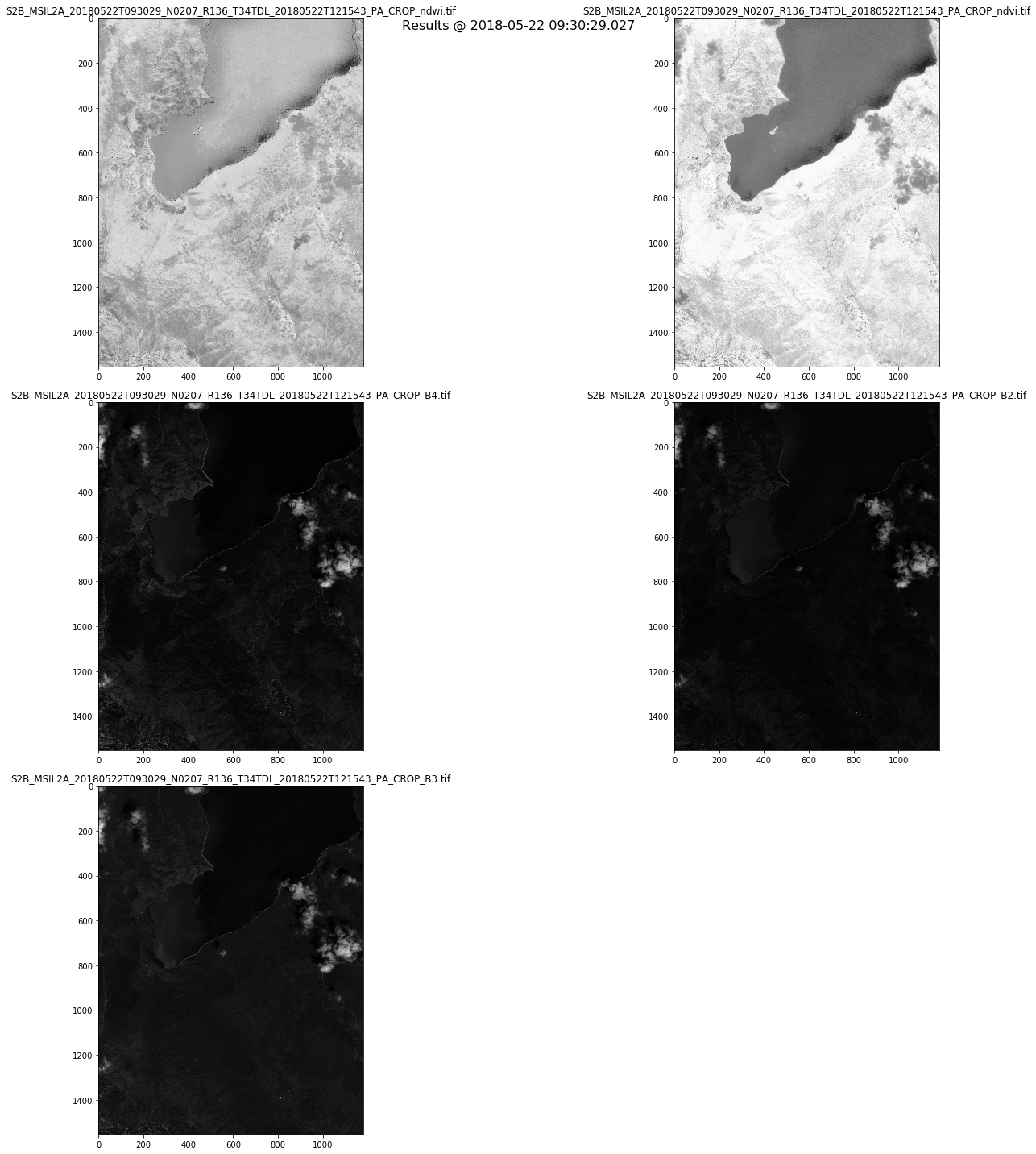
- Plot them as an RGB
In [23]:
rgb_bands = (bands[4],bands[2],bands[3])
rgb_uint8 = np.dstack(rgb_bands).astype(np.uint8)
width = 10
height = 10
plt.figure(figsize=(width, height))
img = Image.fromarray(rgb_uint8)
imgplot = plt.imshow(img)

Export a Geotiff¶
In [24]:
band_number = 3
cols = mydate_results['xsize'].values[0]
rows = mydate_results['ysize'].values[0]
print (cols,rows)
geo_transform = mydate_results['geo_transform'].values[0]
projection = mydate_results['projection'].values[0]
drv = gdal.GetDriverByName('GTiff')
ds = drv.Create('export.tif', cols, rows, band_number, gdal.GDT_Float32)
ds.SetGeoTransform(geo_transform)
ds.SetProjection(projection)
ds.GetRasterBand(1).WriteArray(bands[0], 0, 0)
ds.GetRasterBand(2).WriteArray(bands[1], 0, 0)
ds.GetRasterBand(3).WriteArray(bands[2], 0, 0)
ds.FlushCache()
(1181, 1555)
Download functionalities¶
Download a product¶
- Define the download function
In [25]:
def get_product(url, dest, api_key):
request_headers = {'X-JFrog-Art-Api': api_key}
r = requests.get(url, headers=request_headers)
open(dest, 'wb').write(r.content)
return r.status_code
- Get the reference download endpoint for the product related to the first date
In [26]:
enclosure = mydate_results[(mydate_results['startdate'] == mydates[0])]['enclosure'].values[0]
enclosure
Out[26]:
'https://store.terradue.com/better-satcen-00001/_results/workflows/ec_better_ewf_satcen_01_01_01_ewf_satcen_01_01_01_0_13/run/95ac10ac-315d-11e9-a8e6-0242ac11000f/0012261-181221095105003-oozie-oozi-W/b1a47f4d-ed2a-46cb-95c2-9d32287eb7dd/S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_ndwi.tif'
In [27]:
output_name = mydate_results[(mydate_results['startdate'] == mydates[0])]['title'].values[0]
output_name
Out[27]:
'S2B_MSIL2A_20180522T093029_N0207_R136_T34TDL_20180522T121543_PA_CROP_ndwi.tif'
In [28]:
get_product(enclosure,
output_name,
api_key)
Out[28]:
200
Bulk Download¶
- Define the bulk download function
In [29]:
def get_product_bulk(row, api_key):
return get_product(row['enclosure'],
row['title'],
api_key)
- Download all the products related to a chosen date
In [30]:
date_results = results[(results['startdate'] == mydates[0])]
date_results.apply(lambda row: get_product_bulk(row, api_key), axis=1)
Out[30]:
0 200
1 200
2 200
3 200
4 200
5 200
6 200
7 200
8 200
9 200
10 200
11 200
12 200
13 200
14 200
15 200
16 200
17 200
18 200
19 200
20 200
21 200
22 200
23 200
24 200
25 200
26 200
27 200
28 200
29 200
...
75 200
76 200
77 200
78 200
79 200
80 200
81 200
82 200
83 200
84 200
85 200
86 200
87 200
88 200
89 200
90 200
91 200
92 200
93 200
94 200
95 200
96 200
97 200
98 200
99 200
100 200
101 200
102 200
103 200
104 200
Length: 105, dtype: int64